Where do HS rat genotypes have gaps?
- By: Faith Okamoto
- Original development: March 2023
- Writeup: July 2024
Introduction
Palmer Lab uses heterogenous stock (HS) rats to investigate the genetics of various traits. Phenotypes and genotypes are analyzed in tandem to find potentially relevant regions of the genome. However, this only works if the genotypes include those regions. Sometimes a signal appears but is then "cut off" by a lack of genotype data. In other cases, the signal may be missed entirely if that region is not genotyped.
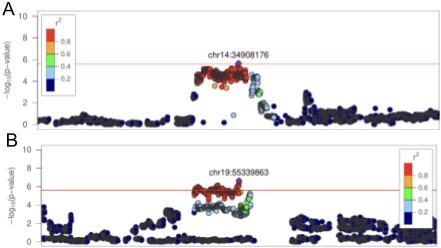
Figure 1. Example plots, from Gunturkun et al. 2022. A. Ideal case. Regional association plot for "SIT: Total travel distance". Variant position on X-axis, variant association significance on Y-axis. Note the peak in the middle, surrounded by variants with low significance. Taken from Supplementary Figure S49. B. Problematic case. Regional association plot for "SIT: Total distance to social zone". Note the peak in the middle, bordered by a region with no genotyped variants. Taken from Supplementary Figure S47.
There are many possible causes for such a "gap" in genotyped variants. The region may be intrinsically difficult to genotype (e.g. many repeats). A lack of genetic variation is also possible. The founders of the HS rat population were chosen to maximize genetic diversity. However, with only 8 strains included, the amount of variation is finite (Hansen and Spuhler 1984). Also, due to breeding a finite number of rats for ~100 generations, genetic drift caused even more loss of genetic variation.
It is of interest to know the locations and causes of HS rat genotype gaps. For example, little can be done to address a gap due to a lack of founder variation, but a gap due to poor genotyping may be amenable to improved genotyping methods. This project set out to discover gap locations in the context of minor allele frequency (MAF), both in founders and the modern population.
Materials & Methods
Datasets
Founders were genotyped with ~40x coverage by whole-genome sequencing (WGS) of one male from each of the 8 founder strains. Single nucleotide polymorphisms (SNPs) and indels on mRatBN7.2 were called by GATK, as described in Chen 2022. Variants were filtered by missing rate (maximum 10%; with only 8 founders, this removes all missingness). The final dataset had 11,191,459 variants called in 8 rats.
Modern HS rats were genotyped with ~0.25x coverage by double-digest genotyping by sequencing (Gileta et al. 2020) or low-coverage WGS. Biallelic SNPs on mRatBN7.2 were imputed by STITCH, as described in Chen et al. 2023. SNPs were filtered by missing rate of at most 0.1 as well as obedience to Hardy-Weinberg equilibrium (p-value of at least 10-10). Hardy-Weinberg equilibrium tests skipped for Y Chromosome (chrY) variants, and performed among only females for X Chromosome variants. The final dataset had 7,069,124 SNPs called in 19,117 rats.
Note that the mitochondrial chromosome was omitted. It is quite small (16kbp) and has well-called variants. For a detailed analysis of HS rat mitochondria, see Okamoto et al. 2023.
Gap finding
PLINK1.9's --freq flag (applied to non-MAF-filtered genotypes) was used to determine each variant's MAF. SNP IDs were assumed to have the format <chr>:<pos>. Gap distance between variants was calculated by subtracting neighboring variant positions (within each chromosome), and then filtering for a minimum meaningful gap length (10kb).
To find gaps created by MAF filtering (i.e. a lack of genetic diversity, as opposed to a lack of well-genotyped markers), this process was repeated while ignoring variants with MAF greater than 0.005, the threshold Palmer Lab uses in standard analyses.
Software
- PLINK version
1.90b6.21 64-bit (19 Oct 2020), used for calculating allele frequencies - R version
4.3.3, used for plotting results. Packages used:data.tableversion1.15.4ggplot2version3.5.1
Code is in GitHub (requires access to Palmer Lab's GitHub account).
Results/Discussion
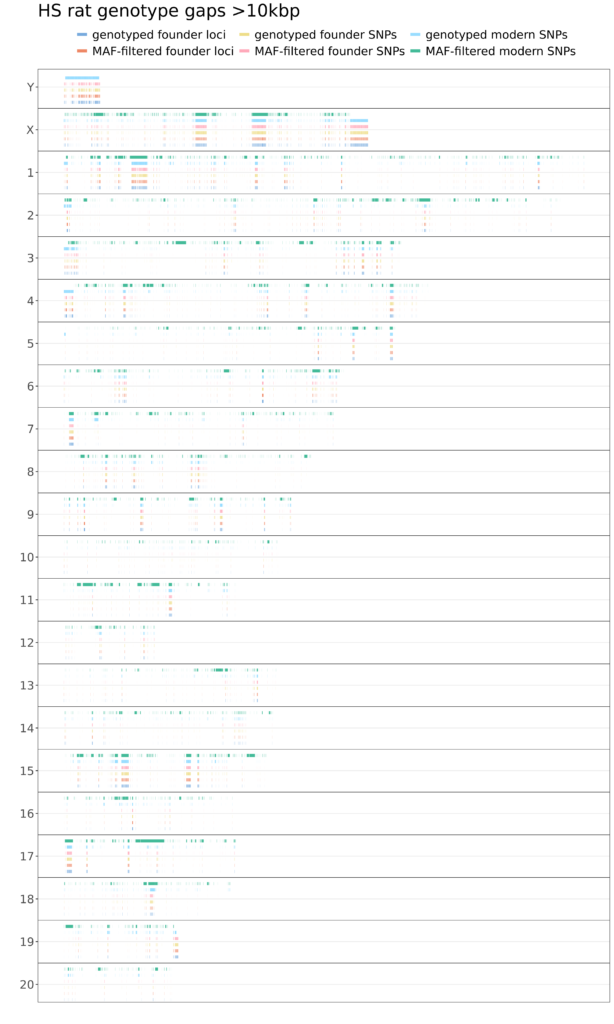
Figure 2. HS rat genotype gaps. Chromosome separated in lines. Variant position along chromosomem on X-axis. Colored line indicates gap of at least 10kb between genotyped variants.
Large gaps are distributed over various chromosomes, significantly increasing after MAF filtering is applied. Some notable feaures include:
- Many gaps in chrY, except there are no gaps in modern genotypes after MAF filtering. This is due to loss of all chrY SNPs: no SNPs remain to have a gap between them! That a weakness of the standard genotyping pipeline, likely due to half of the rats having high missingness on chrY. For a detailed analysis of HS rat chrY, see Okamoto et al. 2023.
- Quite large gaps in chrX and chr1. Indeed, after MAF filtering, there are no variants at the end of chrX at all.
- Large gaps in chr11 and chr17 which appear only after MAF filtration, and then only in the modern (Round 10) rats. This indicates that significant genetic variation has been lost in these region.
- Smaller gaps dot all chromosomes and all regions, only sometimes due to a lack of variation in founders or modern rats. Thus, there is room to improve the variant set and genotyping pipeline to reduce the number and size of gaps.
- There are a few smaller gaps which appear in the founder genotype data when filtering out indels. However, no major gaps are due to sparse SNPs where indels exist. Indels are not currently called in the low-coverage modern HS rat data.
References
Chen D. 2022. Palmer Lab High Coverage WGS Genotyping Pipeline. doi:10.5281/zenodo.6584834.
Chen D, Chitre A, Cheng R, Peng B, Polesskaya O, Palmer A. 2023. Palmer Lab Heterogeneous Stock Rats Genotyping Pipeline. doi:10.5281/zenodo.10002191.
Gileta AF, Gao J, Chitre AS, Bimschleger HV, St. Pierre CL, Gopalakrishnan S, Palmer AA. 2020. Adapting Genotyping-by-Sequencing and Variant Calling for Heterogeneous Stock Rats. G3 Genes|Genomes|Genetics. 10(7):2195–2205. doi:10.1534/g3.120.401325.
Gunturkun MH, Wang T, Chitre AS, Garcia Martinez A, Holl K, St. Pierre C, Bimschleger H, Gao J, Cheng R, Polesskaya O, et al. 2022. Genome-Wide Association Study on Three Behaviors Tested in an Open Field in Heterogeneous Stock Rats Identifies Multiple Loci Implicated in Psychiatric Disorders. Front Psychiatry. 13. doi:10.3389/fpsyt.2022.790566.
Hansen C, Spuhler K. 1984. Development of the National Institutes of Health Genetically Heterogeneous Rat Stock. Alcohol: Clinical and Experimental Research. 8(5):477–479. doi:10.1111/j.1530-0277.1984.tb05706.x.
Okamoto F, Chitre AS, Sanches TM, Chen D, Munro D, Rats NC for G in O, Polesskaya O, Palmer AA. 2023. Y and Mitochondrial Chromosomes in the Heterogeneous Stock Rat Population. :2023.11.29.566473. doi:10.1101/2023.11.29.566473. [accessed 2024 Jul 8].



Comments are closed.