Detecting errors by using albinism
- By: Faith Okamoto
- Original development: April 2023
- Writeup: July 2024
Introduction
Palmer Lab uses phenotypes and genotypes collected from heterogenous stock (HS) rats. All data is tied to a rat's RFID, but is collected separately. Typically a collaborator collects phenotypes (e.g. weighs the rat) and records those values in a spreadsheet. Then we extract DNA and add barcodes, which are recorded in another spreadsheet. Finally, sequenced reads are separated by barcode. The final genotypes thus have an RFID attached to each sample for cross-reference with phenotypes.
As the (simplified) description suggests, there are many steps with possible errors. For example, two weights may be swapped in the records. Or, the location/barcode of a DNA sample may be mis-recorded. Such "sample mix-ups" effectively randomize the assignment of phenotypes to genotypes. Even a few mix-ups can have terrible consequences for genetic association studies.
Quality control (QC) steps are used to ensure quality genotypes. However, only one addresses sample mix-ups: a "sex QC" compares genetic sex (measured by X/Y Chromosome reads) against phenotyped sex. This is practical as sex is easy to both genotype and phenotype.
This project seeks to introduce and verify a new kind of QC: an "albinism QC". Coat color is easy to phenotype. Also, the genetic basis of albinism in rats is known: it arises in rat homozygous for the missense mutation Arg299His, a single nucleotide polymorphism (SNP) in the Tyr gene, at chr1:141201010 in mRatBN7.2 (finding SNP: Blaszczyk et al. 2005; finding universality: Kuramoto et al. 2012). Of HS rats' eight inbred founder strains (ACI/N, BN/SsN, BUF/N, F344/N, M520/N, MR/N, WKY/N, WN/N; Hansen and Spuhler 1984), this SNP is present in six. Many modern HS rats are albino. An albinism QC compares phenotyped coat color (albino vs. not albino) to genotyped albinism (homozygous recessive vs. others).
Materials & Methods
Dataset
Modern HS rats were genotyped with ~0.25x coverage via double-digest genotyping by sequencing (Gileta et al. 2020) or low-coverage whole-genome sequencing (WGS). Biallelic single nucleotide polymorphisms (SNPs) on mRatBN7.2 chr1 were imputed by STITCH, as described in Chen et al. 2023. Post-imputation, rats were filtered to those with a phenotyped coat color, and SNPs were filtered to those in a 1Mbp window centered on chr1:141201010. SNPs were then filtered by standard thresholds:
- minor allele frequency (MAF) of ≥ 0.005
- missing rate of ≤ 0.1
- Hardy-Weinberg equilibrium p-value of ≥ 10-10
The final dataset had 3,430 SNPs (including the causal SNP) and 17,025 rats. Coat color was recorded at phenotyping centers as one of albino (2,925), black (3,589), brown (3,484), black hood (3,584), or brown hood (3,443).
The low-coverage data was also, separately, subset to the causal SNP without filtering out rats which lacked a phenotyped coat color.
A subset of 88 HS rats underwent high-coverage WGS. SNPs and indels on mRatBN7.2 were called by GATK, as described in Chen 2022. These genotypes were filtered to just the causal SNP.
Association testing
Normal statistical tests (e.g. chi-squared test for recessive model) do not work well for scanning SNP associations with albinism. The p-values are too small for e.g. R to compute. Thus, a simpler approach was used: count putative errors. For each SNP, alleles were assigned albino/non-albino to minimize error under a recessive model. Error was calculated as the sum total of homozygous recessive non-albinos and non-homozygous-recessive albinos.
Software
- PLINK version
1.90b6.21 64-bit (19 Oct 2020), used for genotype filtration - R version
4.2.3, used for data analysis. Packages used:
Code for QC and figures is available in GitHub (Palmer lab only).
Results/Discussion
High-coverage genotypes for causal SNP match coat color
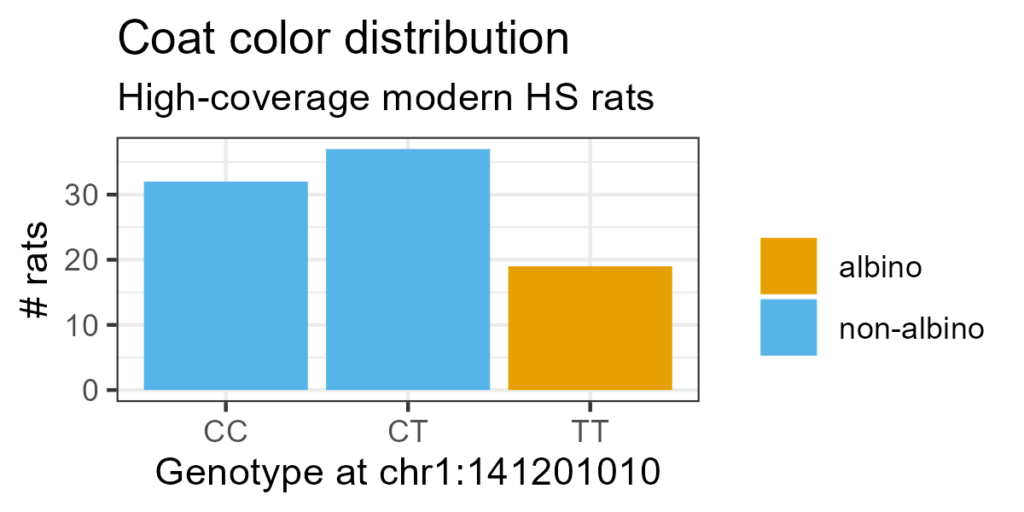
Figure 1. Distribution of coat colors by genotype at causal SNP. Genotype on X-axis, number of rats on Y-axis. Bars colored by coat color.
The causal SNP behaves as expected in the 88 modern HS rats with high-coverage, high-confidence genotypes. Albinism follows an autosomal recessive pattern.
Albinism SNP is a reasonably good predictor of albinism phenotype
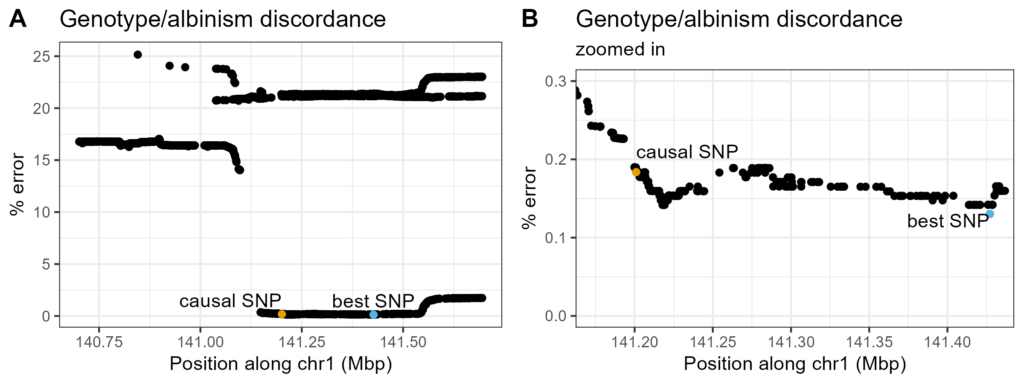
Figure 2. Errors from running a hypothetical albinism QC for SNPs near causal one. Position along chr1 on X-axis, percent error of match to phenotyped coat color on Y-axis. Causal SNP and SNP with best percent error picked out in different colors. A. SNPs in a 1Mbp window centered on causal SNP. B. Panel A zoomed in to area around causal and best SNP.
To ensure low-coverage genotyping/imputation works well for the causal SNP, an albinism QC was run for SNPs in a 1Mbp window around it. The causal SNP has a quite low percent error, though not the best; the best SNP (chr1:141427166, 0.13% error) is nearby. This may be due to random genotyping error/missingness around the causal SNP. While a "better" result is achieved by using a non-causal SNP, the causal SNP is good enough for the actual QC.
Development of albinism QC
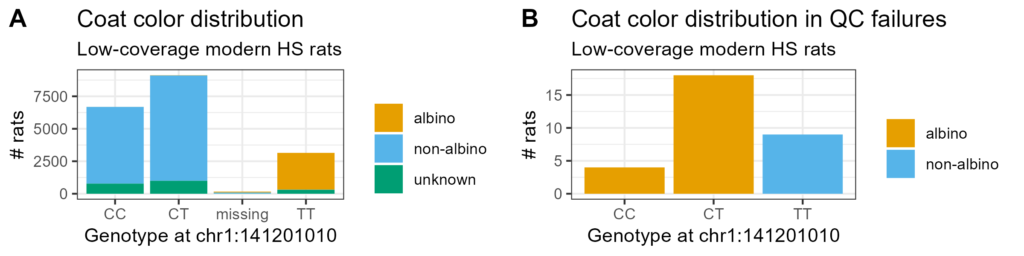
Figure 3. Distribution of coat colors by genotype at causal SNP. Genotype on X-axis, number of rats on Y-axis. Bars colored by coat color. A. All rats with low-coverage data, including those missing genotype/phenotype for the QC. B. Only rats which failed the QC.
Some rats could not undergo albinism QC. Of rats genotyped with low coverage, 2,092 had no recorded coat color (these were ignored in the previous section), and 161 (including 13 with no coat color) had no genotype for the causal SNP.
Among the remaining rats, albinism QC showed high concordance between genotype at the causal SNP and phenotyped coat color. 31 failed the QC and 16,846 passed (failure rate: 0.18%). Note that not all sample mix-ups would be caught, if rats were swapped within albinism groups. Genotyping may have some non-negligible error rate due to the low-coverage genotyping methods used. Still, these 31 rats likely warrant further investigation as possible sample mix-ups.
Conclusion
Albinism QC is both simple to implement and has power to detect putative sample mix-ups. It is thus useful to add to Palmer Lab's standard genotyping QC.
References
Blaszczyk WM, Arning L, Hoffmann K-P, Epplen JT. 2005. A Tyrosinase missense mutation causes albinism in the Wistar rat. Pigment Cell Research. 18(2):144–145. doi:10.1111/j.1600-0749.2005.00227.x.
Chen D. 2022. Palmer Lab High Coverage WGS Genotyping Pipeline. doi:10.5281/zenodo.6584834.
Chen D, Chitre A, Cheng R, Peng B, Polesskaya O, Palmer A. 2023. Palmer Lab Heterogeneous Stock Rats Genotyping Pipeline. doi:10.5281/zenodo.10002191.
Gileta AF, Gao J, Chitre AS, Bimschleger HV, St. Pierre CL, Gopalakrishnan S, Palmer AA. 2020. Adapting Genotyping-by-Sequencing and Variant Calling for Heterogeneous Stock Rats. G3 Genes|Genomes|Genetics. 10(7):2195–2205. doi:10.1534/g3.120.401325.
Hansen C, Spuhler K. 1984. Development of the National Institutes of Health Genetically Heterogeneous Rat Stock. Alcohol: Clinical and Experimental Research. 8(5):477–479. doi:10.1111/j.1530-0277.1984.tb05706.x.
Kuramoto T, Nakanishi S, Ochiai M, Nakagama H, Voigt B, Serikawa T. 2012. Origins of Albino and Hooded Rats: Implications from Molecular Genetic Analysis across Modern Laboratory Rat Strains. PLOS ONE. 7(8):e43059. doi:10.1371/journal.pone.0043059.



Comments are closed.